About NCBI API Key
Who needs to get a key?
For most casual use, you won’t need an API key at all – you only need to get one if you expect to access E-utilities at a rate of more than three requests per second from a single computer (IP address). Even for higher rates of usage, you won’t need a key before May 1, 2018. After May 1, 2018, any computer (IP address) that submits more than three E-utility requests per second will receive an error message. This limit applies to any combination of requests to EInfo, ESearch, ESummary, EFetch, ELink, EPost, ESpell, and EGquery. Just to be clear, this rate applies only to when we receive the request, not to how long the request executes. You can have more than three concurrent requests running as long as you don’t initiate more than three requests in any one-second window. Even though you won’t need a key before May 1, keys are already available. You can get one now and begin using it right away.
As NCBI mentioned, do not send more than three concurrent requests per second, but you can have more than three concurrent requests running.
For example:
1
2
3
4
# Generate a list of 5 numbers
echo -e "1\n2\n3\n4\n5" | \
# Use parallel to run your command 5 times concurrently
parallel -j 3 'esearch -db genome -query "Streptococcus pseudopneumoniae[Organism]" | efetch -format docsum'
Executing above code will concurrently send 3 requests to NCBI, and it will return results normally. If the job number is 4:
1
2
3
4
# Generate a list of 5 numbers
echo -e "1\n2\n3\n4\n5" | \
# Use parallel to run your command 5 times concurrently
parallel -j 4 'esearch -db genome -query "Streptococcus pseudopneumoniae[Organism]" | efetch -format docsum'
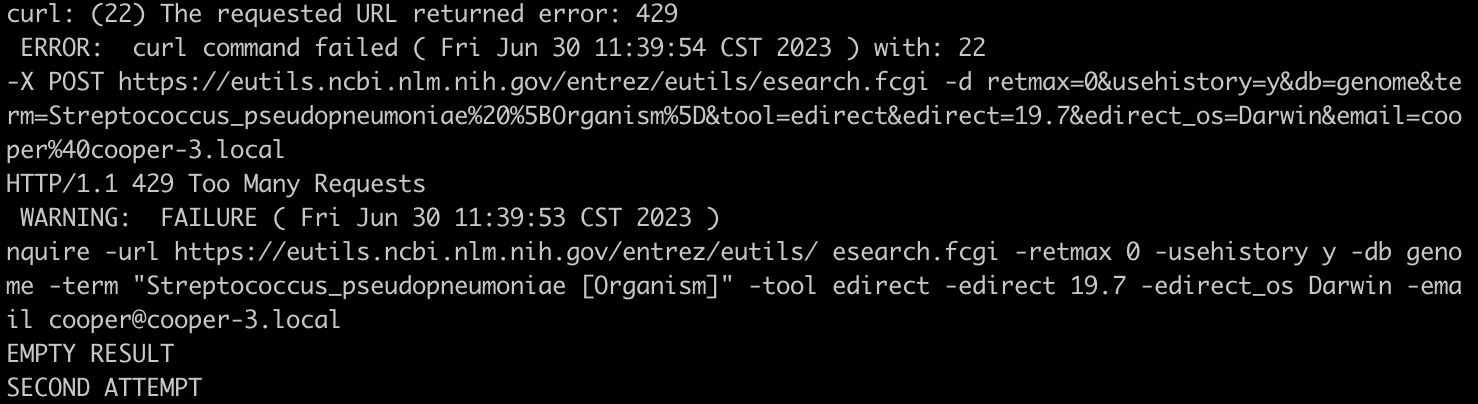
One of the requests will receive HTTP error 429, which is NCBI is telling you “Too many requests”.
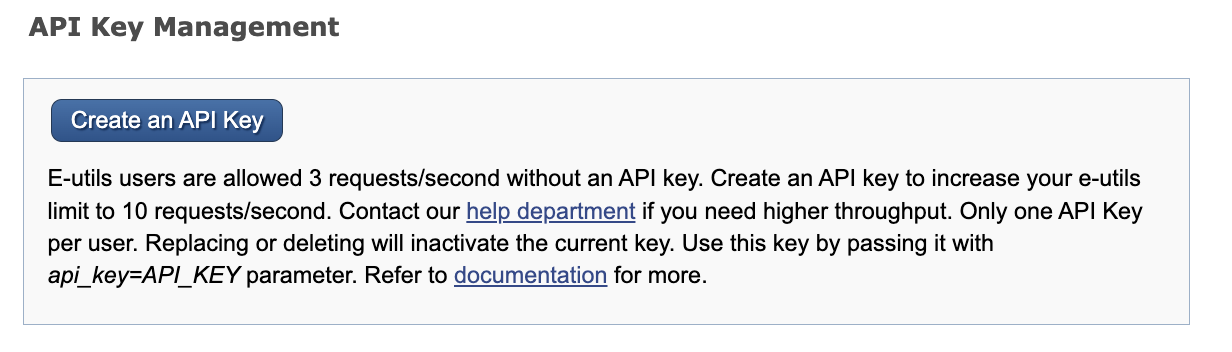
After having an API key, the limit is up to 10 requests per second. Then, set your API key in the terminal:
1
export NCBI_API_KEY=93e327fe6b734742035f50587eb4fc12ae08
“93e327fe6b734742035f50587eb4fc12ae08” is my key, replace it with yours.
1
2
3
4
# Generate a list of 5 numbers
echo -e "1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n11\n12" | \
# Use parallel to run your command 5 times concurrently
parallel -j 12 'esearch -db genome -query "Streptococcus pseudopneumoniae[Organism]" | efetch -format docsum' |grep "Error"
If you attempt to send 12 requests concurrently, you might encounter 2 error messages. However, this isn’t a consistent occurrence; there are instances where the system handles more than 10 requests per second without any issues.
An instance: Assuming you want to download a bacteria’s all assembly genomes
1
esearch -db assembly -query "Streptococcus pseudopneumoniae[Organism] AND latest[filter] NOT anomalous[filter]" |efetch -format docsum | xtract -pattern DocumentSummary -element FtpPath_RefSeq > download.url
the above download.url file looks like this:
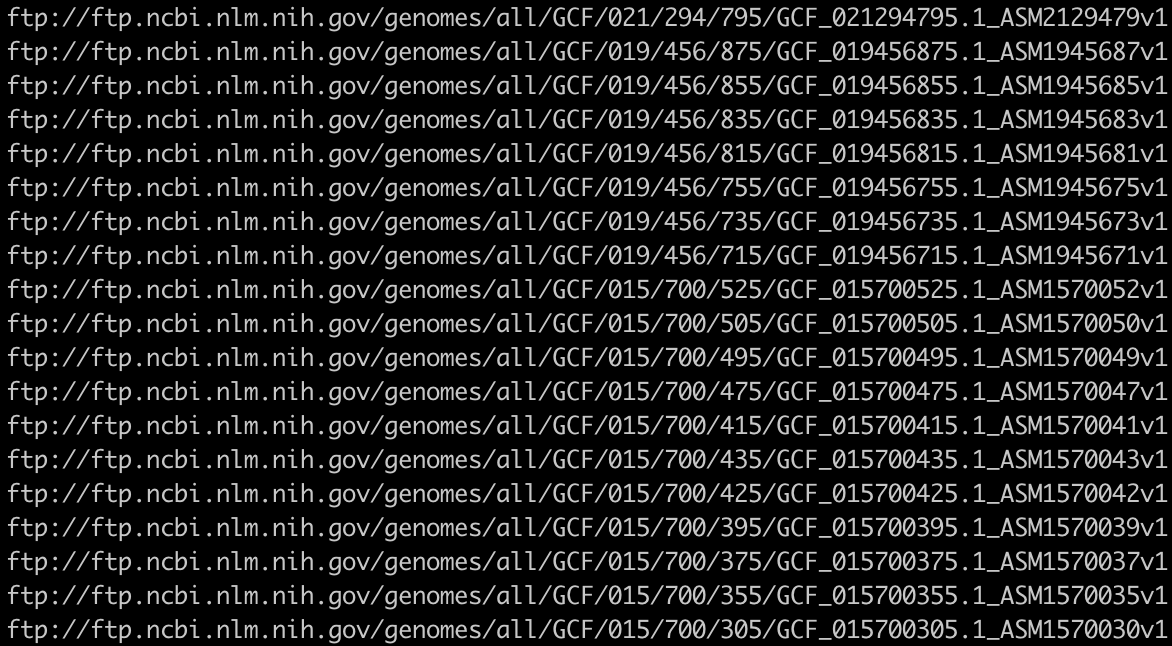
You can use wget to download those file, for example:
1
wget -e use_proxy=yes -e http_proxy=http://127.0.0.1:8001 -r ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/076/775/GCF_001076775.1_ASM107677v1
Even wget is using an http proxy, the speed is still slow. It’s obvious that the file is hosted by an ` ftp service, maybe the reason is the inconsonance between http and ftp`.
If you replace the “ftp:” with “http:” of each url, and open it with browser, you will see this:
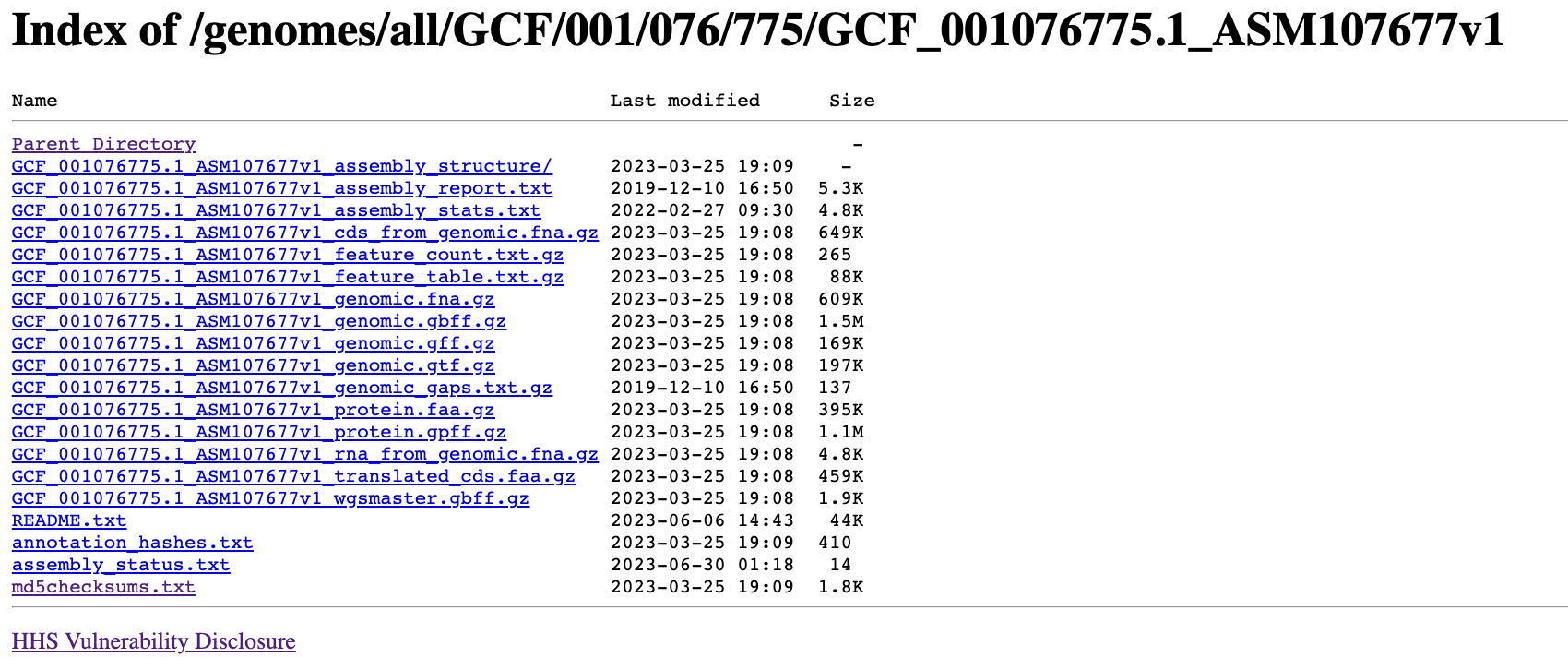
It seems NCBI also host those file behind an http server with Nginx.
Then, I try to use curl instead of wget to download files from NCBI.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
#!/bin/bash
# check if a file name has been provided as a parameter
if [ -z "$1" ]
then
echo "Please provide a file name as a parameter."
exit 1
fi
# read each line from the file
while IFS= read -r line
do
# replace ftp: with https:
url=${line/ftp:/https:}
# extract the last part of the URL
base=$(basename "$url")
# add the "_genomic.fna.gz" string to the end of each url
url+="/${base}_genomic.fna.gz"
# add the url to the urls array
urls+=("$url")
echo $url
# curl -O $url
done < "$1"
# export a function for parallel to use
download_url() {
curl -O "$1"
}
export -f download_url
# download files concurrently using parallel
printf "%s\n" "${urls[@]}" | parallel -j12 download_url
==In a word, you can use `curl` to concurrently download files from NCBI with no limit.== However, the true limit is your network bandwidth and CPU resources.
#E-utilities vs E-direct
E-direct is part of E-utilities